Article Text
Abstract
Objective The remodelling of gut mycobiome (ie, fungi) during pregnancy and its potential influence on host metabolism and pregnancy health remains largely unexplored. Here, we aim to examine the characteristics of gut fungi in pregnant women, and reveal the associations between gut mycobiome, host metabolome and pregnancy health.
Design Based on a prospective birth cohort in central China (2017 to 2020): Tongji-Huaxi-Shuangliu Birth Cohort, we included 4800 participants who had available ITS2 sequencing data, dietary information and clinical records during their pregnancy. Additionally, we established a subcohort of 1059 participants, which included 514 women who gave birth to preterm, low birthweight or macrosomia infants, as well as 545 randomly selected controls. In this subcohort, a total of 750, 748 and 709 participants had ITS2 sequencing data, 16S sequencing data and serum metabolome data available, respectively, across all trimesters.
Results The composition of gut fungi changes dramatically from early to late pregnancy, exhibiting a greater degree of variability and individuality compared with changes observed in gut bacteria. The multiomics data provide a landscape of the networks among gut mycobiome, biological functionality, serum metabolites and pregnancy health, pinpointing the link between Mucor and adverse pregnancy outcomes. The prepregnancy overweight status is a key factor influencing both gut mycobiome compositional alteration and the pattern of metabolic remodelling during pregnancy.
Conclusion This study provides a landscape of gut mycobiome dynamics during pregnancy and its relationship with host metabolism and pregnancy health, which lays the foundation of the future gut mycobiome investigation for healthy pregnancy.
- INTESTINAL MICROBIOLOGY
Data availability statement
Data are available in a public, open access repository. The raw data of ITS2, 16S and metagenomic sequencing in this study have been deposited in the Genome Sequence Archive (GSA) (https://ngdc.cncb.ac.cn/gsa/) at accession number CRA014764; CRA014766; CRA014529. Analysis R codes, Stata codes as well as the annotation pipelines for gut fungi, gut bacteria and gut microbial function are available via https://github.com/nutrition-westlake/THSBC-gut_fungi.Other data sets generated during and/or analysed during the current study are available from the corresponding author upon reasonable request.
This is an open access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited, appropriate credit is given, any changes made indicated, and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/.
Statistics from Altmetric.com
WHAT IS ALREADY KNOWN ON THIS SUBJECT
The human intestine is home to a diverse range of bacterial and fungal species, forming the ecological community that contributes to normal physiology.
The human gut ecological community changes during pregnancy and plays a role in gestational dysmetabolic conditions.
The remodelling of gut fungi during pregnancy and its potential influence on host metabolism and pregnancy health remains largely unexplored.
WHAT THIS STUDY ADDS
The compositional changes of gut fungi from early to late pregnancy exhibit a greater degree of variability and individuality compared with changes observed in gut bacteria.
The prepregnancy overweight status is a key factor influencing both gut mycobiome compositional alteration as well as the pattern of metabolic remodelling during pregnancy.
Gut fungal Mucor during early pregnancy is positively associated with the risk of gestational diabetes mellitus and macrosomia.
The multiomics data provide a landscape of the networks among gut fungi, biological functionality, serum metabolites and pregnancy health.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Our findings indicate the dynamic nature of the gut mycobiome throughout each trimester of pregnancy and its impacts on host metabolism as well as pregnancy health.
The manipulation of the gut fungi, a crucial constituent of the gut ecological community, holds great potential to serve as a novel approach to promoting a healthy pregnancy.
Introduction
During normal pregnancy, the maternal body undergoes dramatic physiological changes including immunological, hormonal and metabolic changes.1 2 Gut microbiota is considered as a virtual organ, and pregnancy status is associated with a profound alteration of the gut microbiota.3 4 It is hypothesised that the host can manipulate the gut microbiota to promote metabolic changes during pregnancy, ultimately supporting the growth and development of the fetus.4
It is important to note that the gastrointestinal tract is populated not only by bacteria, but also by fungi, known as gut mycobiome.5–7 However, fungal communities are still far less studied, compared with the extensive research conducted on the gut bacteria. As a major component of the gut microbiota, gut fungi are believed to play a crucial role in intestinal ecology, which is essential for host health.8 9 Recently, several studies have investigated the gut mycobiome in pregnant women, particularly among those who are obese or have been diagnosed with gestational diabetes mellitus (GDM).10–12 Although the sample sizes of these studies are limited, they provide crucial indications that the gut mycobiome may interact with host metabolism during pregnancy and influence the development of GDM. Therefore, the gut mycobiome has the potential to be an intervention target for promoting a healthy pregnancy.
Regarding the remodelling of gut mycobiome during pregnancy, however, the dynamics of gut mycobiome and its interactions with gut microbial functionality, host metabolism, pregnancy complications and adverse birth outcomes have not been well studied. Here, we aim to comprehensively examine the underlying determinants for gut mycobiome based on a large-scale cohort of pregnant women (n=4800) and profile the dynamics of gut mycobiome based on a subcohort of deeply phenotyped participants (n=750). Moreover, leveraging the repeat measurements of multiomics and deep phenotypes in the established subcohort, we aim to provide a landscape of the networks among the gut mycobiome, gut microbial functionality, host metabolism and pregnancy health.
Results
Participant characteristics and gut mycobiome composition
This study was based on a prospective birth cohort study in central China: Tongji-Huaxi-Shuangliu Birth Cohort (THSBC). The THSBC recruited pregnant women who initiated prenatal care in a local maternal and child health hospital during their early pregnancy. Exclusion criteria were (1) Receiving infertility treatment (eg, in vitro fertilisation or intrauterine insemination); (2) Reporting severe chronic or infectious diseases (eg, cancer, HIV infection or tuberculosis); or (3) Were unable to or refused to sign the informed consent. In the present analysis, we included 4800 participants who had available ITS2 sequencing data, dietary information and clinical records during their pregnancy. This data set enables us to comprehensively profile the gut mycobiome among pregnant women and investigate potential determinants contributing to the variations of the gut mycobiome. To examine how pregnancy impacts the gut mycobiome over time and investigate their potential associations with host metabolism, we established a subcohort of 1059 participants, which included 514 women who gave birth to preterm (n=240), low birthweight (n=137) or macrosomia (n=216) infants, as well as 545 randomly selected participants who did not experience the above three adverse pregnancy outcomes. ITS2 sequencing was performed for the whole cohort involving 4800 participants, while the shotgun metagenomics sequencing was performed for the first trimester (T1) samples within the established subcohort (n=1059). Additionally, within the subcohort, 750 and 748 participants had ITS2 and 16S sequencing data available, respectively, across all trimesters. Figure 1 provides an overview of the study workflow.
Study workflow for profiling the gut fungi during pregnancy and exploring its relationship with host metabolism and health. To comprehensively profile the gut mycobiome-host interaction among pregnant women, we investigated potential determinants contributing to the variations of the gut mycobiome and explored the impact of gut mycobiome during early pregnancy on later pregnancy complications as well as birth outcomes in a large cohort involving 4800 pregnant women. To examine how pregnancy impacts the gut mycobiome over time and investigate potential associations between the gut mycobiome and host metabolism, we established a subcohort of 1059 participants, which included 514 women who gave birth to preterm (n=240), low birth weight (n=137) or macrosomia (n=216) infants, as well as 545 randomly selected healthy controls. Within this subcohort, ITS2 sequencing was performed on 1059 stool samples collected during the first trimester of pregnancy, 890 during the second trimester of pregnancy and 850 during the third trimester. A total of 750 participants in this subcohort had ITS2 sequencing data available for all trimesters.
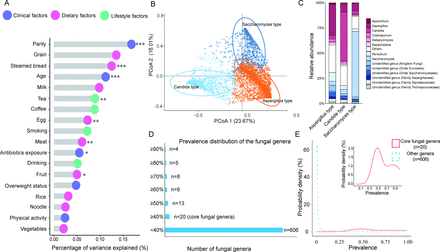
The age of the included participants (n=4800) ranged from 18 years to 40 years (mean age, 26.4; SD, 3.6; table 1). More than half (57.5%) of these women were primiparous, while a majority of the remaining participants (41.3%) were multiparous. Prior to pregnancy, 947 women in the study were underweight and 666 women were overweight or obese, while most of the women had normal body weights (n=3187). Age and parity are most important prepregnancy anthropometric factors contributing to the interindividual variation of gut mycobiome composition. Antibiotics use and dietary factors including steamed bread, egg, fruits, meat and tea consumption are also identified as significant contributors (figure 2A, p<0.05).
Characteristics of the study population
Profiling of the gut mycobiome composition and enterotypes among pregnant women. (A) Variance in the mycobiome composition explained by potential determinants was assessed through permutational multivariate analysis of variance (PERMANOVA) analysis. This analysis was performed based on 4800 independent samples collected during the first trimester. The value of p was determined through 999 permutations. Significance levels are indicated as follows: *, p<0.05; **, p<0.01; ***, p<0.001.(B) Clustering results of fungal enterotypes were visualised by principal coordinate analysis (PCoA). This visualisation was applied for all samples collected in the whole cohort. (C) The most abundant genera within each enterotype were shown. This analysis was based on all samples collected in the whole cohort. (D) The number of gut fungal genera that survived prevalence-based filtering at various cut-off thresholds was shown. This analysis was performed based on 4800 independent samples collected during the first trimester. (E) The distribution of prevalence of gut fungal genera was demonstrated. This analysis was performed based on 4800 independent samples collected during the first trimester.
We observed three fungal enterotypes, including Saccharomyces-dominated enterotype (prevalence, 26.5%), Candida-dominated enterotype (18.8%) and Aspergillus-dominated enterotype (54.7%, figure 2B,C). Parity and dietary factors including steamed bread and tea consumption are most important factors influencing the fungal enterotypes (online supplemental table S1). Specifically, women with a Saccharomyces-dominated enterotype during early pregnancy are more likely to be primiparous and have a dietary preference for consuming steamed bread within the past year. On the other hand, women with a Candida-dominated enterotype are more likely to have a lifestyle characterised by tea consumption within the past year. Of note, Saccharomyces, Candida and Aspergillus are the top three prevalent fungi, while the overall prevalence of gut mycobiome is very sparse (figure 2D,E). Specifically, we identify a total of 626 fungal genera at T1 (n=4800), and more than 96% (606 out of 626) of the identified genera have a prevalence lower than 40%. We consider those present among more than 40% of participants as core fungal genera (n=20) in subsequent analyses.
Compositional dynamics of the gut mycobiome during pregnancy in the longitudinal subcohort
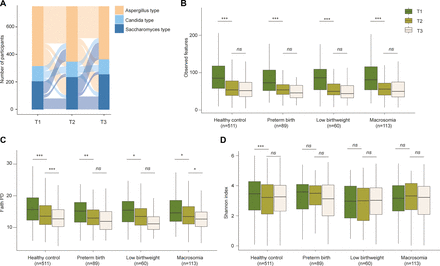
We examined the compositional changes during pregnancy among the 750 participants who had ITS2 sequencing data available for all trimesters. The results showed a global shift in microbial community composition from T1 to the second trimester (T2), but not T2 to the third trimester (T3) (online supplemental figure 1A,B). Inconsistent with prior knowledge that the gut mycobiome was relatively stable,9-10 our findings showed that as many as 68.5% of the 750 participants experienced shifts of the fungal enterotype during pregnancy (figure 3A). Specifically, the proportion of Saccharomyces-dominated enterotype increased (T1, 27.7%; T2, 31.7% and T3, 34.4%) and the Aspergillus-dominated enterotype decreased (T1, 53.1%; T2, 49.3% and T3, 48.4%) from T1 to T3. The proportion of Candida-dominated enterotype was relatively stable throughout the pregnancy (ranging from 17.2% to 19.1%).
Supplemental material
The shifts of gut fungal enterotypes and the dynamics of gut fungal α diversity during pregnancy. (A) Sankey diagram illustrating the shifts of gut fungal enterotypes from early pregnancy to late pregnancy. (B–D) The comparison of gut fungal α diversity across different trimesters, demonstrating the dynamics of gut fungal α diversity from early pregnancy to late pregnancy. Box plot centres show medians of the α diversity metrics with boxes indicating their IQRs, upper and lower whiskers indicating 1.5 times the IQR from above the upper quartile and below the lower quartile, respectively. Paired t test was performed to determine the significance of difference. Significance levels are indicated as follows: ns, p>0.05; *, p<0.05; **, p<0.01; ***, p<0.001. T1, the first trimester of pregnancy; T2, the second trimester of pregnancy; T3, the third trimester of pregnancy.
The within-sample α diversity, including phylogenetic diversity and richness, was substantially reduced from T1 to T3, while the Shannon Index was not that dynamic (figure 3B–D). Moreover, the alterations in the within-sample α diversity during pregnancy were very similar between women who gave birth to healthy infants and those who had preterm, low birthweight or macrosomia infants. Although dietary factors contributed to the gut mycobiome compositional variation across participants, we did not observe significant associations between changes in the consumption of eight main food groups from T1 to T3 and decreased richness, after multiple testing correction (false discovery rate (FDR)>0.05, online supplemental table S2). Therefore, the decreased richness was not likely to be driven by the change of diet during pregnancy.
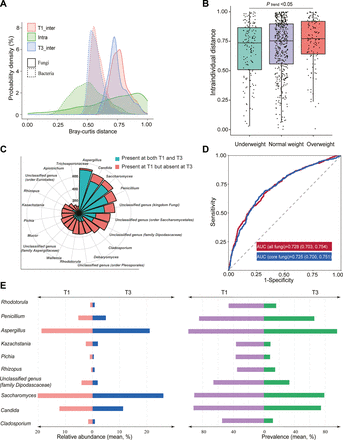
We further profiled the extent of intraindividual shift in the composition in the context of interindividual variation. There was a wide range of intraindividual Bray-Curtis distance from T1 to T2 or T1 to T3, most of which were even larger than the mean interindividual distance at a single time point (figure 4A, online supplemental figure 1C). By contrast, the gut bacteria composition was much more conserved during pregnancy. We observed consistency between compositional alterations and the transition of enterotypes, as participants who exhibited a changed fungal enterotypes displayed considerably greater alterations in gut mycobiome composition (online supplemental figure 1D). Moreover, we found that women who were overweight or obese prior to pregnancy experienced much more variation of gut fungi from T1 to T3 compared with those who were underweight before pregnancy (figure 4B).
Discriminative gut fungal genera between early and late pregnancy. (A) The distributions of variation in gut fungal and bacterial composition over time (from T1 to T3) within individuals, as well as the differences between individuals at T1 or T3. (B) Comparison of the extent of gut mycobiome compositional alteration within individuals over time (from T1 to T3) stratified by prepregnancy overweight status. (C) Nightingale rose diagram visualising the proportion of participants whose core gut fungal genera were lost during later pregnancy in comparison to early pregnancy. (D) A machine learning framework, specifically LightGBM, was employed to train the trimester classifier on the gut mycobiome composition at T1 and T3. Subsequently, this trained classifier was used to predict the trimester to which the samples belong, employing a 10-fold cross-validation strategy and the corresponding area under the curve (AUC) values were presented. (E) The figure displays the relative abundance (left) and prevalence (right) of the top 10 gut fungal genera that contributed to the trimester classifier for T1 and T3.
Dynamics of the individual gut fungal genus during pregnancy in the longitudinal subcohort
Among the 750 participants with gut mycobiome data available across all trimesters, we identified 410 genera in T1 samples. We then assessed the instability for each genus by calculating the loss rate from T1 to T3. For the 390 less prevalent fungi genera, the mean loss rate was as high as 97.6%, indicating extreme instability. By contrast, the 20 core fungi exhibited a mean loss rate of 55.7%, with the Aspergillus, Candida and Saccharomyces being the most stable fungal genera (figure 4C).
To examine the remodelling of the gut fungi from early to late pregnancy, we found that 4 out of the 20 core genera were substantially altered (paired t test on centered log-ratio (CLR)-transformed data between T1 and T3, FDR<0.05). Specifically, these genera were Aspergillus, Cladosporium, Penicillium and Candida, and all these fungal genera were depleted during late pregnancy compared with early pregnancy, which occurred in 40.4%–57.6% of women. Moreover, we used a machine learning algorithm (Light GBM) to discriminate T1 and T3 samples based on the gut mycobiome composition (10-fold cross validation, area under the curve (AUC)=0.728, figure 4D). Using the interpretable Shapley Additive exPlanations (SHAP) value, we found that the core genera accounted for all the top 10 discriminate fungal genera. Then we only used the core fungal genera for discrimination, and the performance was comparable (AUC=0.725, figure 4D). Most of these top discriminatory genera were over-represented in T1 and belonged mostly to the Saccharomycetales, Eurotiales or Capnodiales order of the Ascomycota (n=8). The relative abundance and prevalence of the top 10 discriminate genera during T1 and T3 were shown in figure 4E. Additionally, we conducted a classification analysis to distinguish between T1 and T2 samples, and between T2 and T3 samples. The AUCs for these comparisons were 0.78 and 0.61, respectively (online supplemental figure 2A,B). Moreover, 8 out of the top 10 fungal genera that contributed significantly to the discrimination between T1 and T2 samples, were also top 10 discriminant genera between T1 and T3 samples. These findings suggest that the gut mycobiome features of T2 and T3 samples are quite similar.
Supplemental material
Key microbial functional pathways and host serum metabolites correlated with gut mycobiome
To examine the potential interaction between gut fungi and bacteria in the context of gut microbial functionality, we annotated biological pathways mostly specific to bacteria based on the paired shotgun metagenomics data for T1 samples (n=1039) within the established subcohort. As the gut mycobiome clusters to well-defined gut fungal enterotypes, we then investigated whether the matrix of pathways recapitulates this structure. The fungal enterotypes significantly contributed to the variations of the pathway matrix (p<0.05, R2=0.69%, online supplemental figure 2C), which may indicate an interaction between the gut mycobiome composition and the gut bacterial functionality. As expected, the bacterial enterotypes, identified following the same procedure as that of the fungal enterotypes, explained much more variations of the pathway matrix (R2=5.15%, online supplemental figure 2D).
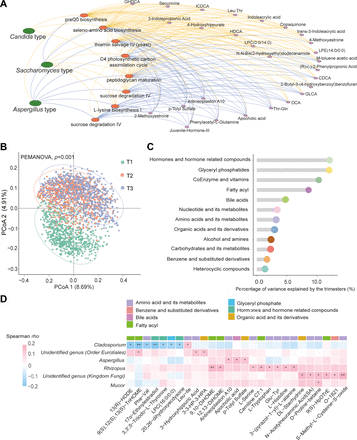
To further characterise the relationship between fungal enterotypes and individual gut microbial functional pathways, we identified eight pathways whose distributions varied across enterotypes (FDR<0.05 with Kruskal-Wallis test). Specifically, most of these pathways (seven out of eight) including thiamin salvage IV (yeast), sucrose degradation IV, L-lysine biosynthesis I, superpathway of L-phenylalanine biosynthesis, peptidoglycan maturation, C4 photosynthetic carbon assimilation cycle and seleno-amino acid biosynthesis, were relatively abundant in the Saccharomyces-dominated enterotype, but less abundant in the Aspergillus-dominated enterotype. At the same time, sucrose degradation IV and seleno-amino acid biosynthesis were also abundant in the Candida-dominated enterotype, while preQ0 biosynthesis was abundant in both Candida-dominated enterotype and Aspergillus-dominated enterotype (figure 5A). Moreover, further regression analysis identified 95 significant associations between the identified pathways and 27 serum metabolites (FDR<0.05, figure 5A). More than two-thirds of the significant metabolites were bile acids (eg, chenodeoxycholic acid, isochodeoxycholic acid, deoxycholic acid, hyodeoxycholic acid and glycolithocholic acid), amino acid metabolites (eg, 4-hydroxyhippurate and phenylacetyl-L-glutamine) and organic acid derivatives. (eg, 3-indolepropionic acid and indoleacrylic acid; figure 5A).
Interactions between gut mycobiome and host metabolism during pregnancy. (A) Network analysis among gut fungal enterotype, microbial functionality and host metabolome. The yellow and blue lines between gut fungal enterotype and functional pathways indicate enrichment and depletion of the pathways, respectively. The yellow and blue lines between functional pathways and serum metabolites indicate positive and negative associations, respectively. (B) Comparison on the overall metabolic pattern of serum samples collected during different trimesters of pregnancy. Principal coordinate analysis (PCoA) was employed, using Canberra dissimilarity, to examine the dissimilarities between all samples. Multivariate PERMANOVA analysis was performed to evaluate the extent to which trimester accounted for the variance in the overall metabolic pattern. The value of p was determined based on 999 permutations. (C) The extent of Canberra dissimilarity-based metabolic alterations during pregnancy for different classes of metabolites over time. To quantify these alterations, we assessed the explained variance of each class of metabolites by trimester using multivariate PERMANOVA. (E) Heatmap of covarying relationship between individual core fungal genera and individual serum metabolites from the first trimester to the third trimester. Spearman correlation analysis was performed between the changes in core gut fungi (CLR-transformed) and changes in metabolites. Significance levels are indicated as follows: *, FDR<0.05; **, FDR<0.01. GHDCA, glycohyodeoxycholic acid; ICDCA, isochodeoxycholic acid; HDCA, hyodeoxycholic acid; CDCA, chenodeoxycholic acid; GLCA, glycolithocholic acid, DCA, deoxycholic acid.
As we repeatedly measured the serum metabolome within the established subcohort, we profiled the metabolic alterations and explored its covarying relationship with gut fungi. We quantified 794 identified metabolites, which belonged to diverse biochemical classes, such as amino acids, lipids, nucleotides and carbohydrates. Similar to the gut mycobiome, the serum metabolome also altered dramatically across trimesters, and the alteration between T1 and T2 was much larger than that between T2 and T3 (figure 5B). The proportion of explained variance by trimester ranged from 1.17% to 11.89% for different classes of metabolites, with hormones and its related metabolites ranking at the top (figure 5C). Correlation analyses between individual core fungal genera and individual metabolites showed 30 covarying relationships involving six genera and 27 serum metabolites (FDR<0.05, figure 5D). Four out of the six identified fungi were annotated to the genus level resolution, including Cladosporium, Aspergillus, Rhizopus and Mucor. Specifically, Aspergillus, Rhizopus and Mucor covaried positively with three, eight and one metabolite, respectively, while Cladosporium covaried negatively with seven metabolites. Most of the Rhizopus-related metabolites belong to fatty acyl and the Cladosporium-related metabolites are mainly amino acids or its metabolites.
Similar to the finding that the intraindividual variation of gut mycobiome composition was different across underweight, normal-weight and overweight women, we found that being overweight prior to pregnancy significantly impacted the pattern of metabolic alterations from T1 to T3 (figure 6A). Subsequently, we performed an analysis to identify metabolites that exhibited significant changes dependent on the prepregnancy overweight status. Our findings revealed that 23 specific metabolites (eg, L-Glycine, L-Arginine, hexadecanedioic acid and carnitine) mainly belonging to amino acids and fatty acyl displayed significant alterations exclusively among underweight women during pregnancy (FDR<0.05, figure 6B). In contrast, 24 metabolites (eg, L-Valine, L-Glutamine, 2-Hydroxycinnamic acid and 4-Hydroxybenzyl alcohol) mainly belonging to amino acids and benzene derivatives were found to be significantly altered solely among overweight women (FDR<0.05, figure 6B). Thus, these metabolites likely contributed to the distinct pattern of metabolic changes observed in underweight or overweight women.
Distinctive metabolic changes stratified by prepregnancy overweight status and the clinical significance of the gut mycobiome during pregnancy. (A) Comparison on the overall metabolic dynamics between subgroups. Principal coordinate analysis (PCoA) was employed, using Canberra dissimilarity, to examine the dissimilarities between pregnant women with different prepregnancy overweight status. The value of p was determined based on 999 permutations. (B) Venn plot showing the number of distinctive and common metabolites that changed significantly from the first trimester to the third trimester. In each subgroup stratified by the prepregnancy overweight status, paired t-tests were conducted for each serum metabolite measured at T1 and T3. An FDR<0.05 was considered statistically significant. For those metabolites which significantly changed solely among pregnant women who were underweight or overweight prior to pregnancy, we showed the fold-change and value of p for each metabolite in the volcano plot. The x-axis shows the log2-transforemd fold-changes, and the y axis indicates the -log (base 10) of the FDR values. Red solid lines indicate the threshold of FDR=0.05. Red dots indicate those metabolites significantly increased from T1 to T3, while blue dots indicate metabolites that significantly decreased. Only the metabolites that were characterised with the highest accuracy and could be matched with internal standards were labelled with compound names in this plot. (C) The relationship between core fungal genera and pregnancy complications as well as adverse birth outcomes. Only the statistically significant associations (FDR<0.05) were illustrated in the forest plot. (D) Mediation analysis among the gut fungal genus Mucor, GDM and macrosomia. (E) Curves show LOESS fit for the relative abundance of the identified taxa based on preterm delivery (green) or not (red). The y-axis indicates the relative abundance. 13(R)-HODE, 13R-hydroxy-9Z,11E-octadecadienoic acid; TriHOME, trihydroxyoctadec-10-enoic acid; 3–3-HP-3-HPA, 3-(3-Hydroxyphenyl)−3-hydroxypropanoic acid; DiHOME, dihydroxyoctadec-12-enoic acid; 9(S)-HpOTrE, 9S-hydroperoxy-10E,12Z,15Z-octadecatrienoic acid; O-1821, 7-(3-Hydroxy-2-(3-hydroxy-5-phenylpent-1-enyl)−5-oxocyclopentyl)hept-5-enoic acid. GDM, gestational diabetes mellitus; LOESS, locally weighted regression; PIH, pregnancy-induced hypertension.
Associations of gut mycobiome with pregnancy outcomes
By examining the prospective associations of each core fungal genera during early pregnancy with pregnancy complications in the whole cohort, we found significantly positive associations between Mucor and incident GDM (OR: 1.15, 95% CI 1.06 to 1.26; FDR<0.05), and positive associations of Wallemia with pregnancy-induced hypertension (PIH; OR: 1.16, 95% CI 1.04 to 1.29; p=0.006, FDR=0.12, figure 6C), after adjustment for potential confounders. Moreover, we validated that both GDM and PIH were risk factors for adverse birth outcomes, including macrosomia and preterm delivery. Specifically, GDM was associated with higher risk of macrosomia (OR: 3.08, 95% CI 1.88 to 5.03; FDR<0.05) and preterm birth (OR: 3.15, 95% CI 1.93 to 5.12; p<0.001, figure 6c). PIH was associated with higher risk of low birth weight (OR: 3.73, 95% CI 1.75 to 7.96; FDR<0.05) and preterm birth (OR: 2.57, 95% CI 1.34 to 4.94; FDR<0.05, figure 6C).
Our analysis of the direct relationship between each core fungal genus and adverse birth outcomes in the whole cohort showed that the relative abundance of Mucor was positively associated with the risk of macrosomia (OR: 1.20, 95% CI 1.07 to 1.35; FDR<0.05, figure 6C). We therefore performed a mediation analysis to test whether the effects of Mucor on the fetal overgrowth were mediated by GDM. We found that the gut fungal Mucor during early pregnancy was associated with macrosomia risk independently of GDM, which suggested that the Mucor and GDM might influence the risk of macrosomia through different pathways (figure 6D). We also explored the associations between the extent of gut mycobiome compositional alterations and adverse birth outcomes within the subcohort, which yielded no significant results. This finding may indicate that the shift of overall gut mycobiome composition was a widely shared phenomenon driven by pregnancy, regardless of birth outcomes. However, this did not preclude that the trajectory of some individual gut fungus might be associated with pregnancy health, as we found significant dissimilarity in the trajectory of Mucor between pregnant women delivering preterm and non-preterm infants (FDR<0.05, figure 6E).
Discussion
To the best of our knowledge, this is the first large-scale prospective cohort study characterising the determinants for gut mycobiome and profiling the gut mycobiome dynamics among pregnant women. We validate three of the previously reported gut fungal enterotypes, and the shifts of gut fungal enterotypes are common during pregnancy. Our quantification analysis of compositional alterations throughout the pregnancy supports a much higher dynamic characteristic of gut fungi compared with the gut bacteria, and highlights that prepregnancy overweight status is a significant contributor to the extent of alterations in gut mycobiome composition. Moreover, we perform network analysis among gut mycobiome, biological functionality, serum metabolites and pregnancy health, identifying that Mucor is prospectively associated with both GDM and macrosomia risk.
Our previous work investigated the determinants and stability of gut mycobiome among middle-aged and elderly adults.9 Gut mycobiome composition was temporally stable while modulated by age, long-term habitual diet and host physiological states. A recent study also reported that age could significantly explain the interindividual variation of the human gut mycobiome and strongly affected the fungal enterotypes in several independent cohorts.13–15 In the present study, we found similar significant determinants of gut mycobiome, such as age and diet, but the mycobiome composition was not that stable during pregnancy. The most significant determinants for gut mycobiome among pregnant women included age, parity and dietary intakes of steamed bread, egg, fruits, meat and tea consumption. Moreover, the consumption of steamed bread was related to the Saccharomyces-dominated enterotype while drinking tea was associated with the Candida-dominated enterotype. Interestingly, steamed bread is made by fermenting with Saccharomyces and the fungi are also involved in tea fermentation in China, thus the consumption of steamed bread or fermented tea may directly affect culture-independent gut mycobiome composition.
The present study provides evidence that gut mycobiome composition and structure are unstable and the well-defined gut fungal enterotypes could be altered over the course of the pregnancy. Taking the gut bacteria dynamics during pregnancy as a comparison, the longitudinal intraindividual distance for gut fungi is highly individualised and the mean intraindividual distance is much larger than that for gut bacteria composition. This finding was also true for stool samples collected among Human Microbiome Project volunteers, which showed more similar faecal bacterial community structure than faecal fungal community structure over time.16 The longitudinal samples of one individual’s faecal fungal mycobiome are even less similar to each other than those of another individual. Prior studies had shown differences in maternal gut bacterial composition by prepregnancy weight, indicating considerable effects of prepregnancy body mass index on gut microbiota composition during pregnancy.17–19 In the present study, we report that the prepregnancy overweight status has a substantial influence on the alterations in gut mycobiome composition during pregnancy. These results together highlight the importance of body weight management before pregnancy for maintaining a stable gut microbial ecosystem during pregnancy.
We postulate that the considerable alterations in gut mycobiome composition during pregnancy may be attributed to the extremely high loss rate of the less prevalent fungal genera. The less prevalent (prevalence <40%) fungal genera accounted more than 96% of the identified genera during early pregnancy, and these genera showed an average loss rate of 97.6% from T1 to T3. On the other hand, this finding may also support that most of the identified gut fungi are passengers other than residents during pregnancy. Nevertheless, we identified a group of core taxa for pregnant women in this study and about half of these core genera were also classified as core fungi for middle-aged and elderly individuals in our previous report and were consistently detected in the Human Microbiome Project and Danish cohorts.9 16 Therefore, the identified core fungi are more likely to be resident commensals in the human gastrointestinal tract. This provides a rationale for future research focusing on these prevalent core fungi.
Prior studies have indicated host remodelling of the gut microbiome and metabolic changes during pregnancy, which may potentially impact maternal and infant health.4 20–24 The majority of these studies have primarily focused on gut or vaginal bacteria and metabolites, while very few studies investigated the gut fungi during pregnancy based on a prospective cohort study. Here we present evidence that the interaction between the Saccharomyces-dominant enterotype, gut microbial functionality and host metabolism may be particularly significant. The network analysis demonstrated that the Saccharomyces-dominant enterotype was associated with more gut microbial functionalities compared with other fungal enterotypes. A previous study reported that the enterotype dominated by Candida conferred an increased risk of multiple diseases,15 but we could not correlate the Candida-dominant enterotypes with pregnancy complications or adverse birth outcomes among pregnant women. Nevertheless, we found some specific core fungal genera, which were risk factors for pregnancy complications. The abundances of Mucor and Wallemia during early pregnancy were positively associated with the risk of GDM and PIH, respectively. We also validated the reported adverse effects of both GDM and PIH on birth outcomes, such as macrosomia, preterm birth and low birth weight.25–27 Moreover, the abundance of Mucor during early pregnancy was also directly associated with the risk of macrosomia, which was independent of GDM. These findings collectively highlight that Mucor and Wallemia may serve as examples that support the vital role of gut fungi in impacting pregnancy health.
Although the relationship between Mucor and blood glucose homoeostasis has not been well studied, prior research reported that the Mucor was associated with higher blood glucose levels and inflammatory activity among patients with non-alcoholic fatty liver.28 On the other hand, high blood glucose level is one of the important factors facilitating the growth of Mucor.29 Our findings strengthen the relationship between Mucor and glucose metabolism disorders during pregnancy. Moreover, in a murine model, Mucor administration increased intestinal permeability in epithelial cell monolayers, which might be indicative of unstable intestinal barriers.30 Of note, the increase in the permeability of the gut barrier is thought to contribute to systemic inflammation and diabetes development, and worsen the microvascular complications of existed diabetes.31 32 A prior study investigating the gut mycobiota of patients with GDM from middle to late pregnancy also supports that patients with GDM host a predominance of fungal taxa with potential inflammatory effects.10 Regarding the potential impact of Mucor on fetal overgrowth, there are several species belonging to genus Mucor (eg, Mucor circinelloides) which can provide important alternative sources of bioactive lipids, due to its high efficiency in synthesising and accumulating lipids.33 These findings may help propose potential mechanisms underlying the detrimental effects of Mucor on pregnancy health. Nevertheless, the pathogenic effects of fungi could be species-dependent or even strain-dependent.11 34 Further studies are needed to validate the underlying mechanisms.
Wallemia has been reported to be a member of fungal microbiota in the human gut,35 but it is less well known compared with Candida or Saccharomyces. Wallemia represents one of the most xerophilic fungal taxa, including the most xerophilic, osmophilic, and even halophilic and chaophilic microorganisms described to date.36 37 Several species from the genus Wallemia were reported to produce several bioactive metabolites or toxins, which exhibited antiproliferative and antimicrobial activities.38 39 Previous animal experiments reported that altered Schaedler flora mice colonised with Wallemia mellicola experienced enhanced severity of allergic airways disease compared with fungus-free control mice.40 In the present study, Wallemia was positively associated the risk of PIH, while the underlying mechanisms are yet to be investigated. Nevertheless, Wallemia are found in various osmotically challenged environments, such as dry, salted or highly sugared foods, so we could not rule out that the Wallemia in the present study is just a biomarker of highly salted foods, which may confound our findings.
Our study has several strengths. First, the large sample size enables us to comprehensively profile the gut fungal characteristics and investigate determinants of variations in gut mycobiome composition among pregnant women. Second, the well-established subcohort with longitudinally repeated sample collections and ITS2 sequencing, facilitate the extensive profiling of gut fungal dynamics during pregnancy. Finally, the multiomics data including ITS2 and 16S sequencing, short-gun metagenomics sequencing and serum metabolomics data provide a landscape of the networks among gut fungi, biological functionality and host metabolites. Our study also has several limitations. The pathogenic or probiotic effects of gut fungi could be species-dependent or even strain-dependent, therefore the resolution at the genus level is not high enough in the present study. Future research may improve the accuracy of reference databases for fungal taxon alignment based on metagenomics data. Second, the quantification of gut fungi and bacteria relies on relative abundance measurements, which may introduce unexpected bias into the statistical analysis. Even when data transformation techniques or appropriate methods addressing the compositional nature of the data are employed, the potential bias may still exist. Third, although we perform correlation analysis to gain functional insights of the gut mycobiome, we could not directly annotate the functions specific to gut fungi due to the limited reference databases. In the future, it will be crucial to enhance the function annotation of the human gut mycobiome. Lastly, this study only includes Chinese women, which may inevitably limit the generalisability of our findings.
In summary, we provide evidence for the dynamic nature of gut fungi in comparison to gut bacteria during pregnancy, revealing that prepregnancy overweight status may be a key determinant for this alteration. This study presents a landscape of the networks among the gut mycobiome, biological functionality, serum metabolites and pregnancy health, pinpointing the link between fungal genus Mucor and adverse pregnancy outcomes. This study also provides a reference database and resource for future investigation on the functional role of gut mycobiome in healthy pregnancy.
Data availability statement
Data are available in a public, open access repository. The raw data of ITS2, 16S and metagenomic sequencing in this study have been deposited in the Genome Sequence Archive (GSA) (https://ngdc.cncb.ac.cn/gsa/) at accession number CRA014764; CRA014766; CRA014529. Analysis R codes, Stata codes as well as the annotation pipelines for gut fungi, gut bacteria and gut microbial function are available via https://github.com/nutrition-westlake/THSBC-gut_fungi.Other data sets generated during and/or analysed during the current study are available from the corresponding author upon reasonable request.
Ethics statements
Patient consent for publication
Ethics approval
This study involves human participants and was approved by the Ethics Committee of the Tongji Medical College, Huazhong University of Science and Technology (2017) (S225)-1. Participants gave informed consent to participate in the study before taking part.
Acknowledgments
The authors thank the High-Performance Computing Center and High-Throughput Core Facility at Westlake University for assistance in computing and data generation.
References
Supplementary materials
Supplementary Data
This web only file has been produced by the BMJ Publishing Group from an electronic file supplied by the author(s) and has not been edited for content.
Supplementary Data
This web only file has been produced by the BMJ Publishing Group from an electronic file supplied by the author(s) and has not been edited for content.
Footnotes
YF, WG, PW and YL are joint first authors.
X @zheng_jusheng
YF, WG, PW and YL contributed equally.
Contributors J-SZ, AP, X-FP and YF conceived the study concept and design. YF, WG, PW and YL analysed the data. PW, YL, JY, BZ, YY, XL, YH contributed to the field work, data collection and data curation. YF, KZ, XL, MS, JT, ZM and JC contributed to visualisation of the data. YF and J-SZ wrote the first draft of the manuscript. AP and X-FP contributed to the critical revision of the manuscript. YF, WG, PW and YL contributed equally to the work. All authors approved the final version of the manuscript for publication. J-SZ, AP and X-FP are the guarantors of this work and, as such, had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Funding This study was funded by the National Key R&D Program of China (2022YFA1303900, 2023YFC3606300 and 2022YFC3600600), the National Natural Science Foundation of China (82103826,82073529, U21A20427, 82325043, 81930124, 82192902, 82021005 and 92374112), 'Pioneer' and 'Leading goose' R&D Program of Zhejiang (2022C03102), Zhejiang Provincial Natural Science Foundation of China (LQ21H260002), the Research Program of Westlake Laboratory of Life Sciences and Biomedicine (202208012), the Fundamental Research Funds for the Central Universities (YJ202346), China Postdoctoral Science Foundation (2023M733177, 2022M722833). The funders had no role in collecting data, study design, interpretation of data or the decision to submit the manuscript for publication.
Competing interests None declared.
Patient and public involvement Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review Not commissioned; externally peer reviewed.
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.