Article Text
Abstract
The high genetic diversity of hepatitis C virus (HCV) has led to the emergence of eight genotypes and a large number of subtypes in limited geographical areas. Currently approved pangenotypic DAA regimens have been designed and developed to be effective against the most common subtypes (1a, 1b, 2a, 2b, 2c, 3a, 4a, 5a and 6a). However, large populations living in Africa and Asia, or who have migrated from these regions to industrialised countries, are infected with ‘unusual’, non-epidemic HCV subtypes, including some that are inherently resistant to currently available direct-acting antiviral (DAA) drugs due to the presence of natural polymorphisms at resistance-associated substitution positions. In this review article, we describe the origin and subsequent global spread of HCV genotypes and subtypes, the current global distribution of common and unusual HCV subtypes, the polymorphisms naturally present in the genome sequences of unusual HCV subtypes that may confer inherently reduced susceptibility to DAA drugs and the available data on the response of unusual HCV subtypes to first-line HCV therapy and retreatment. We conclude that the problem of unusual HCV subtypes that are inherently resistant to DAAs and its threat to the global efforts to eliminate viral hepatitis are largely underestimated and warrant vigorous action.
- HCV
- GENOTYPE
- HEPATITIS C
- ANTIVIRAL THERAPY
This is an open access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited, appropriate credit is given, any changes made indicated, and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/.
Statistics from Altmetric.com
Introduction
Hepatitis C virus (HCV) infection is a major global health problem, with approximately 57 million people living with chronic viremic infection worldwide, causing 290 000 deaths per year and 1.5 million new infections annually.1 2 The treatment of hepatitis C has been transformed in recent years by the advent of direct-acting antiviral (DAA) therapy, with definitive cure rates of >95% in clinical trials and in the real world with pangenotypic regimens.3–10 In this context, WHO has set the goal of eliminating hepatitis C as a major public health threat by 2030, with the objective to achieve annual incidence ≤5 per 100 000 and annual mortality ≤2 per 100 000 at country level.11
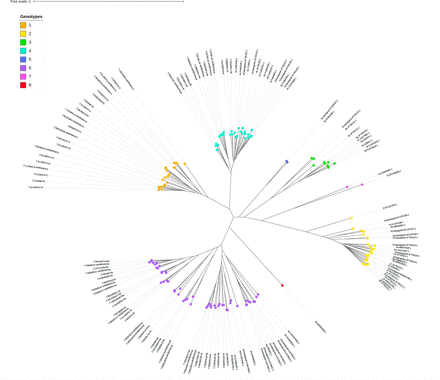
HCV is characterised by high genetic diversity. The Darwinian evolution of HCV has led to the progressive diversification and spread of genotypes and subtypes. Currently, 8 genotypes and over 100 subtypes have been reported, and more may exist (figure 1).12 However, only a small number of HCV subtypes account for the vast majority of infected patients in industrialised countries, including genotypes 1a, 1b, 2a, 2b, 2c, 3a, 4a, 5a and 6a.13 Currently approved pangenotypic DAA regimens have been designed and developed to be effective against these ‘usual’, common HCV subtypes. However, other subtypes, which are considered ‘unusual’ in industrialised areas, have been shown to be prevalent in certain regions of the world and in patients living in Western countries who were born and infected in these regions and subsequently migrated. Lower cure rates than those previously described in Europe and North America have been reported with DAA regimens in patients infected with certain unusual HCV subtypes.14–19 These are due to the presence of natural polymorphisms in the sequence of the viral genome that cause amino acid changes that confer reduced susceptibility to DAAs, principally NS5A inhibitors, making these subtypes inherently resistant to the action of the corresponding DAA.14 20–22
Phylogenetic tree of all known, assigned, provisionally assigned or unassigned subtypes of the eight hepatitis C virus (HCV) genotypes based on genomes from the NCBI Genbank. The genome sequences were aligned using the MAFFT V.7 software, and then the tree was constructed using the maximum likelihood method with IQTree V.2.1.4 based on the GTR+F+I+G4 substitution model (determined automatically by the software). The graphical representation was generated from the iTOL website. The same HCV genome sequences are included in figure 3 and online supplemental table.
Supplemental material
With approximately 30 million patients infected with HCV living in the WHO regions of Africa, Southeast Asia and the Western Pacific,1 the prevalence of unusual HCV subtypes among patients living in these areas and among migrants from these regions, who represent a substantial proportion of newly diagnosed patients with HCV in industrialised countries, may challenge WHO elimination goals. This review article discusses the origin and distribution of unusual HCV genotype subtypes (including non-1a, −1b, −2a, −2b, −2c, −3a, −4a, −5a and −6a), their sensitivity to currently used DAAs and their behaviour during treatment and retreatment with different DAA regimens.
Origin and spread of HCV genotypes and subtypes
Origin of HCV genotypes/subtypes
The genetic variability of HCV results in a typical Darwinian evolutionary process, in which the continuous diversification of viral populations leads to competition between them and to selection of the fittest variants by the environment in which the virus replicates.23 The continuous production of new mutations has led to the permanent selection of variant strains during evolution in geographically or epidemiologically distinct populations, resulting in the emergence and subsequent diversification of the HCV genotypes (phylogenetic clades) and subtypes (phylogenetic subclades) (figure 1).24 The HCV genotypes differ from each other by approximately 30%–35% and the subtypes by >15% of their nucleotide sequence.
HCV genotype/subtype diversification and spread
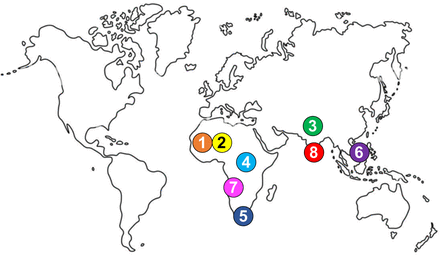
The different genotypes and subtypes of HCV have emerged and been transmitted in restricted geographical areas for hundreds of years: West Africa for genotype 1 and 2 subtypes, Central Africa for genotype 4 subtypes, the Indian subcontinent for genotype 3 subtypes and Southeast Asia for genotype 6 subtypes (no region has yet been found to contain high levels of HCV genotype 5, 7 and 8 genetic diversity) (figure 2).25 26 As a result, residents or migrants from these regions harbour ‘endemic’ HCV subtypes, which are characterised by a low prevalence, long-term local persistence, low transmission rates and a high genetic diversity.27–30
Regions of origin and subsequent diversification of the eight known HCV genotypes (1–8).
In contrast, ‘epidemic’ HCV subtypes (which were originally endemic in their region of origin) spread rapidly during the 20th century through highly effective transmission routes such as blood and blood product transfusion, injection drug use and/or invasive medical procedures.31–33 Epidemic HCV subtypes 1a, 1b, 2a, 2b, 2c, 3a, 4a and, to a lesser extent, 5a and 6a are now globally distributed and cause most HCV infections worldwide. Large-scale public health interventions in the 20th century also led to local epidemics of HCV subtypes. This is the case for subtype 1b in Japan or subtype 4a in Egypt.34–36 Whether different HCV genotypes/subtypes are associated with different clinical outcomes in patients remains controversial, although several studies have suggested that genotype 3 is associated with more severe liver disease.
Not surprisingly, most research interest to date has been directed towards the development of prevention strategies, drugs or vaccines against these highly prevalent epidemic subtypes, as they cause the majority of HCV morbidity in the countries where the studies were conducted.
Determination of HCV genotypes and subtypes
HCV genotyping assays are based on (1) reverse hybridisation of PCR products targeting a specific region of the genome (5’ non-coding region, core region) or (2) sequencing, including population sequencing or next-generation sequencing, of the NS5B region or, less commonly, other genomic regions, followed by phylogenetic analysis.
Reverse hybridisation identifies only the most prevalent, usual HCV subtypes that are predefined in the assay. Therefore, reverse hybridisation is not able to identify unusual HCV subtypes, which are generally assigned to a different usual subtype of their genotype with this method (unpublished personal data).37 38 In contrast, sequencing of the NS5B region accurately identifies the genotype and the subtype, provided that the phylogenetic analysis uses a complete database in which each of the known HCV subtype sequences is represented. Sequencing of the NS5A region also accurately identifies the genotype and the subtype, provided that the database is complete, as well as the presence of resistance-associated substitutions (RASs) in this region.
Distribution of unusual HCV genotypes and subtypes
Worldwide distribution of HCV genotypes and subtypes
The common HCV subtypes are globally distributed. Genotype 1 (1a, 1b), 2 (2a, 2b, 2c) and 3 (3a) subtypes cause the majority of hepatitis C cases worldwide, whereas genotypes 4–8 are less prevalent and more geographically restricted. Globally, genotype 1 (mainly subtypes 1a and 1b) accounts for 46.2% of all HCV cases.39 40 Genotype 3 is found in 30.1% of hepatitis C cases, while genotypes 2, 4 and 6 represent 9.1%, 8.3% and 5.4% of cases, respectively. Genotype 5 is found in <1% of cases.39 40
The true global and local prevalence of unusual HCV subtypes is not well-known, as data are lacking in many countries in Africa and Asia.41 Only 0.8% of patients were African in a global epidemiology study based on the results from phase II and III clinical trials of new DAAs.42 The presence of unusual HCV subtypes can be inferred from patients living in industrialised areas who were infected in their country of origin. However, data are only available from a few Western European countries and none from North America, Northeast Asia or Australia. In addition, molecular epidemiology studies are outdated or suffer from selection bias in many industrialised countries where these subtypes have only recently been introduced. Simplification of HCV therapy without the need for genotype determination prior to treatment initiation has also hindered regular updates of HCV subtype prevalence in many areas of the world.
Distribution of unusual HCV subtypes in Africa
A large diversity of unusual HCV genotype 1 subtypes, including subtypes 1c–1m and unassigned subtypes, is observed in western sub-Saharan Africa, along the Atlantic coast from Senegal to Cameroon, suggesting that this is the region where genotype 1 has emerged and diversified (figure 2).30 43–46 For example, subtypes 1e, 1h or 1l represented 80% of the 108 non-1a/1b HCV genotype 1 subtypes in a study in Cameroon.45 The diversity of unusual genotype 1 subtypes in this region is reflected in immigrants born in these areas and living in industrialised countries. In a recent study at a London hepatology centre, 91 of 2211 patients (4.1%) followed for hepatitis C between 2010 and 2018 were born in Africa, mostly in sub-Saharan Africa. 39% of them were infected with an unusual HCV genotype 1 subtype, including subtypes 1e, 1g, 1h, 1l or an unassigned genotype 1 subtype. These patients were mainly from Benin, Cameroon, Ivory Coast, Ghana or Nigeria.14 Similar data were reported in the HCV Research UK Cohort, a nationwide real-world cohort study from the UK that included 319 patients born in Africa: unusual genotype 1 subtypes (mainly 1c, 1l and unassigned) were found in patients born in West Africa, including Cameroon, Gambia or Nigeria.15
In a nationwide French study, the prevalence of unusual non-1a/1b subtypes of genotype 1 was 0.8% in 2019.47 In our recent study of 640 patients who failed to achieve sustained virological response (SVR) after receiving an NS5A inhibitor-containing regimen analysed at the French National Reference Center for Viral Hepatitis B, C and D, 47 patients (7.3%) were infected with an unusual genotype 1 subtype, including the following: 1d (n=8), 1e (n=13), 1f (n=1), 1g (n=2), 1i (n=2), 1k (n=1), 1l (n=18) and 1 undetermined (n=2). Except for two patients born in France, their countries of origin were Cameroon (n=17), Ivory Coast (n=1), Congo (n=1), Central African Republic (n=1), Togo (n=1), Egypt (n=2), Haiti (n=1) or an unspecified African country (n=14).21
The prevalence and genetic diversity of genotype 2 are also high in West Africa, suggesting that this genotype, like genotype 1, has emerged and diversified in this region, although at a different time (figure 2).48 Indeed, a wide variety of genotype 2 subtypes have been reported in patients from Guinea,49 Guinea-Bissau,50 Ghana,27 51 Ivory Coast,52 Cameroon50 53 and Nigeria.52 Data suggest that the transatlantic slave trade and colonial history have been the driving forces behind the global spread of HCV genotype 2, from West African regions to Central and North African regions54 and from Ghana and Benin to the Caribbean (where genotype 2 further diversified) and then between the Netherlands and its former colonies of Indonesia and Suriname over the past 150 years.55 Genotype 2 is less common and less diverse in Central Africa than in West Africa, suggesting epidemic importation.
The region of origin and diversification of genotype 4 appears to be Central Africa, where a large number of its subtypes circulate in the HCV-infected population (figure 2).56 In Central Africa, genotype 4 subtypes account for more than 80% of HCV infections,57–59 and the most common subtypes are 4c, 4d, 4e, 4k and 4r.56 59–61 Subtypes 4f, 4p, 4t, 4c and 4o (in order of frequency) have been observed in Cameroon53; 4e, 4c, 4f, 4t, 4k, 4r, 4g and unassigned 4 in Gabon57; 4k, 4c, 4r, 4f and unassigned 4 in the Central African Republic62; 4r, 4c, 4k, 4h and unassigned 4 in the Democratic Republic of the Congo60 63; and 4k, 4r, 4q and 4v in Rwanda.18 The prevalence of genotype 4 subtypes is generally unknown in East Africa where data are scarce. In Ethiopia, the majority of HCV strains are genotype 4, with subtypes 4d and 4r accounting for 92%.61 Genotype 4 subtypes, including 4d and 4o, have been identified in Saudi Arabia with a low prevalence.64
Only subtype 5a and one yet unassigned subtype of genotype 5 are known. Subtype 5a is the most common genotype in South Africa, where the majority of cases are found (figure 2).39 65 Genotype 7 (subtypes 7a and 7b) has recently been identified in Congolese immigrants in Canada66 67 and France68 and in Angolan immigrants in South Africa.69
Distribution of unusual HCV subtypes in Asia
In Asia, particularly in Central Asia, East Asia, Southeast Asia and the Asia Pacific region, genotype 1, predominantly subtype 1b, is the most common HCV genotype.70 In a nationwide study in China, the most common subtype was 1b (63.4%), followed by 2a (17.3%), both common subtypes that originated in Africa and subsequently spread throughout the world.71
Endemic Asian HCV genotypes include the following: genotype 3, which emerged, diversified and circulated in South Asia, where it may account for more than 60% of cases in certain regions, such as Pakistan, India or Malaysia70, and genotype 6, which emerged, diversified and circulated in Southeast Asia, where its subtypes predominate in certain regions, such as Laos, Cambodia or Vietnam (figure 2).70 72 Few data are available on the distribution of the different subtypes of genotypes 3 and 6 in Asia, with considerable regional variation.
In an Indian study of people who inject drugs (PWIDs), subtype 3a was found in 39.0% of cases, followed by 1a in 26.9% of cases. In addition, 20.7% of patients were infected with subtype 3b, while other unusual subtypes (1c, 3i, 4d, 6n and 6v) accounted for <5% of cases.73 HCV subtype 3b was more common in individuals co-infected with HIV.74 In Thailand, subtype 3a is predominant (36.4%), followed by 1a (19.9%), 1b (12.6%), 3b (9.7%) and 2a (0.5%). Genotype 3 subtype 3b is more common in the northeastern and southern regions of the country, where it accounts for approximately 15% of cases.75 In a report from Myanmar, subtype 3b was found in nearly 30% of cases.76 In Mainland China, patients infected with subtype 3b represented 54% of those with HCV genotype 3 infection in a national study (compared with <1% in North America or Europe).77 Subtype 3b accounted for 29% of cases among PWIDs from the Yunnan region in another report.78 In Guangdong province, subtype 3b was found in 20.9% of PWIDs and in 3.6% of infected blood donors.79
HCV genotype 6 subtypes circulate mainly in Southeast Asia, including Vietnam (where they have been reported to account for >50% of cases in some studies), Thailand (>30%), Cambodia (>50%), Laos (>90%) and Myanmar (>50%).70 In a study conducted in Thailand, 20.9% of patients were infected with genotype 6, including subtypes 6f (7.8%), 6n (7.7%), 6i (3.4%), 6j and 6m (0.7% each), 6c (0.3%) and 6v and 6xa (0.2% each). The prevalence of genotype 6, mainly subtypes 6f and 6n, was lower in southern Thailand than in the north and northeast of the country (15% vs nearly 30%, respectively).75 In Vietnam, genotype 6 was found in 37.9% of a population of PWIDs, sex workers, patients undergoing dialysis and multiple transfusion recipients, including 6a (18.8%), 6l (6.4%), 6e (6.0%), 6h (4.6%) and unassigned 6 (2.1%).80 In another study, 47% of PWIDs infected with HCV from Yunnan, China, carried genotype 6, including subtypes 6n (30%), 6a (15%), 6u (1%) and 6v (1%).78 In Guangdong province, subtype 6a was found in 57.0% of PWIDs and in 39.8% of infected blood donors. Subtypes 6e and 6k were found in 0.8% and 0.4% of cases, respectively.79
Genotype 6 was found in 36 of 14 603 (0.25%) patients monoinfected with HCV in a French nationwide cohort study. Subtype 6e was the most common (27.8%), followed by 6a (22.2%), 6q (11.1%), 6 unassigned (11.1%), 6o (8.3%), 6p (5.6%) and 6xc, 6f, 6h, 6r and 6t. All but three patients were born in Asia, including Cambodia (44.4%), Vietnam (38.9%) or Laos (8.3%). Subtype 6a was found in 42.8% of Vietnamese and 6e in 37.5% of Cambodian patients, whereas 6q, 6p and 6r were found only in Cambodian patients.81
Recently, a novel genotype 8a was identified in four patients from Punjab, India, living in Canada.82 No other subtypes of genotype 8 are known.
Unusual HCV subtype polymorphisms conferring inherent resistance to HCV DAAs
Quasispecies distribution of HCV populations, natural polymorphisms and resistance-associated substitutions
As previously reviewed,83 the viral populations that constitute the circulating viral quasispecies in the body of infected individuals differ by amino acid polymorphisms that have arisen by mutation during replication and have subsequently been selected based on their impact on viral fitness. Natural polymorphisms located in a viral protein region important for the antiviral activity of a DAA may confer reduced susceptibility to the DAA or DAA class. Such polymorphisms may be present in large, highly fit viral populations or in smaller viral populations if they reduce fitness. When a DAA is administered, positive selection for viral variants with reduced susceptibility to that drug results in viral resistance. The amino acid changes that confer resistance to DAAs are called ‘RASs’, while the viral variants that carry these RASs and thus have reduced susceptibility to the DAA are called ‘resistant variants’.83 Selection of resistant variants carrying RASs explains treatment failure with DAA-based regimens. Compensatory amino acid substitutions, or ‘fitness-associated substitutions’, either naturally present or acquired by mutation during replication of the resistant virus during drug administration, can increase the fitness of resistant variants, leading to their rapid growth on treatment (breakthrough) or after treatment (relapse), and influence their persistence after treatment.84
Commercially available DAAs have been designed to be effective against the most common HCV subtypes (1a, 1b, 2a, 2b, 2c, 3a, 4a, 5a and 6a) and have been clinically validated in northern hemisphere countries where these variants are largely predominant. However, several unusual HCV subtypes common in Africa and Asia and in immigrants from these regions are known to naturally carry NS3 and/or NS5A polymorphisms at RAS positions that confer reduced susceptibility to NS3 protease and/or NS5A inhibitors, respectively. These RASs were initially described after DAA treatment failure in patients infected with common genotypes and subsequently characterised in vitro for drug susceptibility in the genetic context of a common genotype sequence.
The NS3 and/or NS5A RASs are present at baseline as subtype-specific polymorphisms. In the case of treatment failure in patients infected with the corresponding unusual HCV subtypes, the baseline RASs have been selected, while additional RASs that are not naturally present as dominant species, but are co-selected by therapy, are often also present.
RASs carried by unusual HCV subtypes as natural polymorphisms (reference subtype sequences from database)
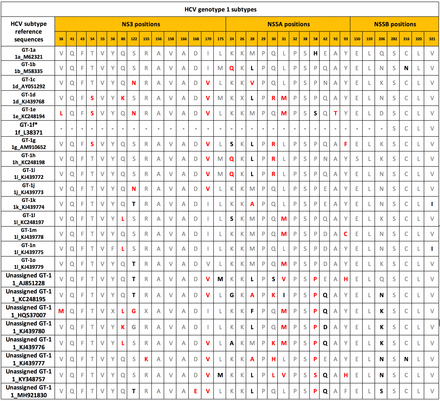
Figure 3 (example of genotype 1 subtypes) and online supplemental table (all HCV genotype subtypes) list the amino acids present at known RAS positions in the NS3 protease, NS5A and NS5B polymerase regions from reference sequences of all known confirmed or yet unassigned HCV subtypes,12 according to the RAS definition in the EASL 2020 Recommendations on Treatment of Hepatitis C, that is, amino acid changes that confer reduced susceptibility to the corresponding drug classes in in vitro assays and/or were selected in patients who failed to achieve SVR on DAA-based regimens.13 Polymorphisms known to confer reduced susceptibility to the corresponding DAAs are shown in bold red, while polymorphisms not associated with reduced susceptibility are shown in bold black. These amino acids are those most commonly found in patients infected with a particular subtype (they may also be present in minority variants in patients infected with common HCV subtypes). However, other amino acids, some of which may also confer reduced susceptibility to DAAs, may be present at specific RAS positions in a minority of patients infected with unusual subtypes.
Most frequent amino acids at known RAS positions in reference sequences from all known assigned, provisionally assigned or unassigned HCV genotype (GT) 1 subtypes (recovered from never treated patients). Amino acids conferring reduced susceptibility to the corresponding DAAs were defined according to the 2020 ‘EASL Recommendations on Treatment of Hepatitis C: Final update of the series’.13 Reference strains were obtained from https://ictv.global/sg_wiki/flaviviridae/hepacivirus/table1.12 Amino acids conferring reduced susceptibility to the DAAs targeting the corresponding regions are shown in bold red. Polymorphisms not reported to be associated with reduced susceptibility to the corresponding DAAs are shown in bold black. Other amino acids are shown in grey. *For these tentatively assigned subtypes, only partial sequences are available in the databases. Other HCV genotype subtypes are shown in online supplemental table. DAA, direct-acting antiviral; HCV, hepatitis C virus; RAS, resistance-associated substitution.
RASs carried by unusual HCV subtypes as natural polymorphisms (patient sequence data)
Several cohort studies have described the presence of polymorphisms conferring reduced DAA susceptibility at baseline in patients infected with different HCV subtypes. The largest included 12 615 patients from 28 countries in 5 geographic regions who were enrolled in industry clinical trials for which sequencing data were available.42 Only a small proportion of the confirmed HCV subtypes were represented in this study, and very few patients infected with the unusual subtypes were enrolled, reflecting the bias towards the inclusion of common subtypes in clinical trials of new DAAs. However, these results, together with those from smaller studies,20 22 77 85–88 confirmed the presence of RASs at baseline in patients infected with certain unusual HCV subtypes.
Polymorphisms at positions K24, M28, Q30, L31, H58, A92 and/or Y93 of the NS5A region have been reported in patients from the above industry study cohort infected with unusual HCV subtypes.42 Certain subtypes naturally harbour combinations of two or three RASs in this region, making them likely to respond poorly to DAA regimens containing an NS5A inhibitor (figure 3, online supplemental table). This was the case for patients infected with unusual genotype 1 subtypes in a recent study of African patients followed at a London centre, who presented with multiple NS5A polymorphisms, particularly at positions 24, 30 and 31. At baseline, RASs conferring reduced susceptibility to NS5A inhibitors or associated with reduced treatment response were present in 82% of these patients.14 Specifically, patients infected with subtype 1l often harbour L31M±Q30 R/L, sometimes associated with other polymorphisms that may play a role in response to DAA therapy.14 21 RASs at baseline are also common in patients infected with unusual genotype 1 subtypes.42
The presence of the A30K RAS is more frequent in non-3a subtypes of genotype 3 than in subtype 3a (84.6% vs 0.8%).89 In addition, the A30K+L31M RAS combination, which confers high levels of resistance to NS5A inhibitors, has been reported in the vast majority of patients infected with unusual subtypes 3b and 3g,16 90 including in 94% of patients infected with subtype 3b in China.91
Pre-existing RASs at multiple NS5A positions, including L28M/V, L30A, L31M and/or Y93H, have been reported in patients infected with subtype 4r, a subtype common in Central Africa and in immigrants from that region.22 87 Various combinations of RASs were observed in eight patients infected with subtype 4r from the above cohort of subjects enrolled in industry clinical trials, including combinations of three NS5A RASs in three of them, two RASs in four of them and one RAS in one of them at baseline.42 In our experience of patients infected with subtype 4r born in Africa and followed in France, 2–3 NS5A polymorphisms were present as dominant species at baseline in all cases, the most common RAS being the double substitution L28V+L30R.20 The Y93H RAS was observed at baseline in 13% of patients infected with subtype 4r but in 50% of those infected with subtype 4b.42 L31M/V/I, which enhances the resistance conferred by Y93H, was present in all patients with subtype 4b and Y93H and in 25% of those with subtype 4r.42
11 of 16 patients (69%) infected with genotype 6 (mainly 6n) had the double F28L+T93S NS5A RAS, although it remains unclear whether F28L actually reduces susceptibility to NS5A inhibitors.92 In another study, R30S was found in 100% of patients infected with subtype 6e and F/L28V in 100% of patients infected with subtypes 6h, 6k and 6l.93
NS3 protease RASs have been identified as natural polymorphisms in three confirmed subtypes of genotype 1, including subtype 1c (R155D, A156T and D168E), subtype 1e (V36L, T54S and S122N) and subtype 1l (Q80L).42 NS3 protease RASs V26L, Q80L and S122R have been reported in subtypes 2i and 2j, V36L and D168Q in subtypes 3b and 3i and V36L and S122T in subtypes 4c, 4k, 4n and 4o.42 In our experience, D168E was found in the dominant viral population of one patient with subtype 4r infection who had never been previously exposed to an NS3 protease inhibitor.20 Q80K and D168E were also detected in subtype 5a (100% and 53%, respectively) and in subtype 6a (100% and 7%, respectively), whereas S122T was observed in subtypes 6e and 6m.42 No NS5B RASs were reported at baseline in patients infected with unusual subtypes.
Little information is available from patients infected with most of the other assigned and unassigned unusual HCV subtypes, which have rarely been sequenced. Based on figure 3, online supplemental table and anecdotal reports, it is likely that many of them carry natural polymorphisms that confer reduced susceptibility to DAAs.
DAA resistance conferred by natural polymorphisms at RAS positions in unusual HCV subtypes in vitro
The degree of DAA resistance conferred in the context of the full-length unusual HCV subtype sequence of the viral protein has generally not been characterised in vitro. One study measured the susceptibility to various NS5A inhibitors of NS5A sequences from some unusual HCV subtypes (including 1l, 3b, 3g, 4r, 6u and 6v) inserted into a subtype 2a replicon backbone. Although the behaviour of infecting strains in vivo may differ from that observed in vitro, the data suggest that subtypes 3b and 3g are resistant to the four NS5A inhibitors daclatasvir, ledipasvir, elbasvir and velpatasvir. Subtypes 1l, 4r, 6u and 6v may be susceptible to elbasvir and velpatasvir but not to daclatasvir and ledipasvir. Only pibrentasvir had high activity against all subtypes tested.94 In another study, RASs observed at baseline in genotype 3 subtypes of patients enrolled in a clinical trial were inserted into a genotype 3a replicon. Polymorphisms at RAS positions commonly found in subtypes 3b and 3g conferred reduced susceptibility to daclatasvir, elbasvir, velpatasvir and pibrentasvir.90 Other subtypes have not been tested in vitro.
Response of unusual HCV subtypes to first-line DAA therapy
Because clinical trials primarily enrolled patients from limited regions infected with the most common HCV genotype subtypes, while the genotyping techniques used generally failed to identify patients infected with unusual subtypes, very little, fragmented information is available on the response to different generations of DAAs of most HCV subtypes, including those carrying baseline RASs that are likely to be poorly responsive to antiviral drugs. In addition, most studies involving patients infected with unusual HCV subtypes used earlier-generation DAA combinations, so little is known about the effect of last-generation pangenotypic first-line regimens (sofosbuvir/velpatasvir and glecaprevir/pibrentasvir) on these subtypes. What patient information is available is summarised below.
HCV genotype 1
In a cohort of 2211 patients with chronic hepatitis C followed at a London centre between 2010 and 2018, 91 patients (4.1%) were born in Africa, mostly in sub-Saharan Africa.14 Of these, 39% were infected with an unusual genotype 1 subtype. 28 of them were treated with various regimens, including sofosbuvir/ledipasvir in 15 cases. They included 2 patients with subtype 1e, 5 with 1g, 3 with 1l, 2 with 1p and 16 with an unassigned subtype. Their SVR rate was only 21/28 (75%), compared with 100% in patients infected with subtypes 1a and 1b. The seven SVR failures were three 1l, one 1p, two unassigned with the combination of sofosbuvir and ledipasvir and one unassigned with the combination of grazoprevir and elbasvir. These patients had RASs at baseline, which conferred reduced susceptibility to the NS5A inhibitor used, which were also present at failure.14
In the HCV Research UK cohort, a nationwide cohort study of 319 patients with HCV infection born in Africa and infected with different HCV subtypes, 53 patients infected with genotype 1 were treated. The SVR rates were 88% (23/26) for subtype 1a and 94% (16/17) for subtype 1b but only 30% (3/10) for subtype 1l. The seven patients infected with subtype 1l who did not achieve SVR had received sofosbuvir plus daclatasvir or sofosbuvir/ledipasvir.15
Between January 2015 and December 2021, we analysed samples collected at the time of relapse in 640 patients who failed to achieve SVR after receiving an NS5A inhibitor-containing regimen at the French National Reference Center for Viral Hepatitis B, C and D. Of these, 47 patients (7.3%) were infected with an unusual genotype 1 subtype, the vast majority of whom were born in Africa.21 This prevalence is much higher than that of unusual genotype 1 subtypes in the French population of patients infected with HCV, which was estimated at 0.8% in a nationwide multicentre study,47 confirming that these patients are more likely to fail to achieve SVR on DAA therapy than those infected with other viral subtypes. Treatment failure occurred with sofosbuvir/ledipasvir in 74.4% of cases, other earlier generation combinations in 14.0% of cases, sofosbuvir/velpatasvir in 4.7% of cases and glecaprevir/pibrentasvir in 7.0% of cases.21
In a German study using the European Resistance Database, which includes samples from patients in several Western and Central European countries, the prevalence of unusual HCV subtypes was 1.6% (75 of 4,653) in DAA-naïve patients and 4.4% (60 of 1,376) in patients who failed to achieve SVR after DAA-based therapy.95 Six patients infected with genotype 1 (1c, n=1; 1e, n=2; 1l, n=1; unassigned 1, n=2) failed DAA therapy, including ombitasvir/paritaprevir/ritonavir plus dasabuvir in one case, grazoprevir/elbasvir in one case and sofosbuvir/ledipasvir in four cases.95 In contrast, the SVR rate was 100% in patients from a Dutch cohort of unusual HCV subtypes infected with subtypes 1c (n=7), 1d (n=1) and 1g (n=10) receiving a pre-pangenotypic regimen in 14 cases, sofosbuvir/velpatasvir in 2 cases and glecaprevir/pibrentasvir in 2 cases.96
HCV genotype 2
The latter Dutch study included 66 patients infected with unusual HCV genotype 2 subtypes out of 160 patients with an unusual subtype treated with DAAs (41%), an exceptionally high prevalence compared with other European series, probably reflecting specificities in the regions of origin of individuals immigrating to the Netherlands. Their distribution was 2c (n=3), 2e (n=3), 2f (n=9), 2i (n=9), 2k (n=4), 2o (n=1), 2p (n=2) and unassigned 2 (n=35). The SVR rate in this group was 93%, with one failure with subtype 2f, one with subtype 2i and two with unassigned genotype 2 subtype. The failures occurred after sofosbuvir plus ribavirin in three cases and sofosbuvir plus daclatasvir in one case.96 These results suggest that HCV genotype 2 subtypes, including unusual ones, are easy to cure with DAAs, pending additional information.
HCV genotype 3
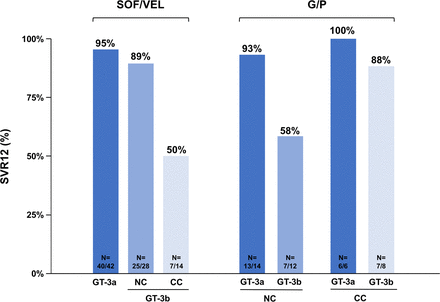
A few clinical trials and real-world studies have been conducted in China in patients infected with HCV subtype 3b, as recently reviewed.97 Globally, SVR rates were lower in patients infected with subtype 3b than in those infected with subtype 3a, including with the latest pangenotypic combinations (figure 4): 76% (32/42) vs 95% (40/42) with sofosbuvir/velpatasvir,16 70% (14/20) vs 95% (19/20) with glecaprevir/pibrentasvir17 and 89% (24/27) vs 91% (21/23) with sofosbuvir plus coblopasvir (an NS5A inhibitor not available outside of China).98 In patients with subtype 3b receiving sofosbuvir/velpatasvir, the SVR rate was 89% (25/28) in those without cirrhosis, but only 50% (7/14) in those with compensated cirrhosis.16 The SVR rate was 58% (7/12) in cirrhotic patients with subtype 3b receiving glecaprevir/pibrentasvir.17
Sustained virological response (SVR) rates in Asian patients without cirrhosis (NC) or with compensated cirrhosis (CC) infected with hepatitis C virus (HCV) subtype 3a (GT-3a) vs subtype 3b (GT-3b) in a single-arm, open-label phase III study (left)16 and in two multicentre phase III studies, the double-blind VOYAGE-1 study and the open-label, single-arm VOYAGE-2 study (right).17 SOF, sofosbuvir; VEL, velpatasvir; G, glecaprevir; P, pibrentasvir.
In the Dutch cohort, three out of eight patients infected with subtype 3b did not achieve SVR. The three failures were with sofosbuvir plus daclatasvir and sofosbuvir/velpatasvir. All three patients infected with subtype 3k achieved SVR.96 In the European Resistance Database study, 15 patients infected with an unusual genotype 3 subtype failed DAA therapy (3b, n=8; 3g, n=2; 3i, n=1; 3h, n=2; 3k, n=1; and unassigned, n=1), including sofosbuvir and daclatasvir in 6 cases, grazoprevir/elbasvir in 1 case, sofosbuvir/ledipasvir in 1 case, sofosbuvir/velpatasvir in 4 cases and glecaprevir/pibrentasvir in 3 cases.95
HCV genotype 4
Of 573 patients infected with HCV genotype 4 included in 18 clinical trials of approved DAA regimens, 49% were infected with subtype 4a, 31% with subtype 4d and only 16% with 1 of 14 other genotype 4 subtypes.99 12 of them experienced virological failure: 7 infected with subtype 4d, 2 with 4r, 1 with 4a, 1 with 4b and 1 with an unassigned subtype. Failures were observed with sofosbuvir/ledipasvir, grazoprevir/elbasvir and ombitasvir/paritaprevir/ritonavir.99 In a pivotal clinical trial of sofosbuvir/ledipasvir in 44 patients infected with genotype 4, 3 subjects did not achieve SVR, including 2 infected with subtype 4r (out of 3 included) and the patient with subtype 4b.87 In the HCV Research UK cohort, which included patients infected with HCV born in Africa and living in England infected with different HCV subtypes, 93% (25/27) of patients were infected with subtype 4a and 100% (4/4) of those infected with subtype 4d, but only 60% (9/15) of those infected with subtype 4r achieved SVR. Most virological failures occurred with sofosbuvir/ledipasvir or sofosbuvir and daclatasvir.15
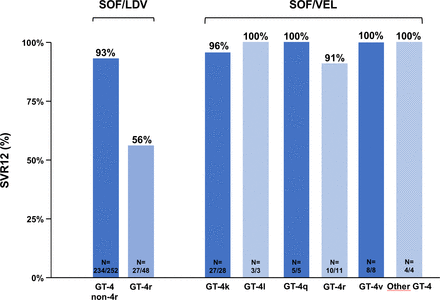
We recently reported our experience at the French National Reference Center for Viral Hepatitis B, C and D of a large over-representation of patients infected with HCV subtype 4r among those who failed to achieve SVR after DAA-based therapy in France, as compared with the very low prevalence of this subtype in the country. Indeed, of 537 patients treated with a DAA-containing regimen who experienced a virological failure, 121 (22.5%) were infected with genotype 4, and 27 of these (22.3%) were infected with subtype 4r.20 These results are in keeping with a clinical trial conducted in Rwanda, in which the 300 patients enrolled were infected with a subtype of genotype 4: 4k in 45% of cases, 4r in 16%, 4q in 14%, 4v in 8%, others (4a, 4b, 4c, 4d, 4g, 4l and mixed) in 10% and unassigned in 7%.18 After 12 weeks of sofosbuvir/ledipasvir, the SVR rate was only 56% (27/48) in patients infected with subtype 4r, significantly lower than the 93% (234/252) SVR rate observed for the other genotype 4 subtypes (figure 5).18 Another study from Rwanda included 58 patients infected with genotype 4 (4k, n=28; 4r, n=11; 4v, n=8; 4q, n=5; 4l, n=3; 4b, n=1; 4c, n=1; unassigned, n=1; and undetermined, n=3) treated with sofosbuvir/velpatasvir. The overall SVR rate was 97% (59/61 patients). 10 of the 11 patients infected with subtype 4r achieved SVR with sofosbuvir/velpatasvir (figure 5).100
Sustained virological response (SVR) rates in patients from Rwanda infected with different subtypes of hepatitis C virus (HCV) genotype 4 (GT-4) treated with sofosbuvir/ledipasvir (left)18 or with sofosbuvir/velpatasvir (right)100 in two single-arm studies. SOF, sofosbuvir; LDV, ledipasvir; VEL, velpatasvir.
These results are consistent with those of the European Resistance Database study, in which 28 patients infected with an unusual genotype 4 subtype failed DAA therapy, including 17 infected with subtype 4r and 11 with other subtypes (4b, 4c, 4f, 4n, 4o, 4v and unassigned 4). Virological failure occurred after treatment with sofosbuvir/ledipasvir in 43% of cases, ombitasvir/paritaprevir/ritonavir in 21% of cases, sofosbuvir and daclatasvir in 14% of cases, grazoprevir/elbasvir in 7% of cases, sofosbuvir/velpatasvir in 7% of cases, sofosbuvir and ribavirin in 4% of cases and glecaprevir/pibrentasvir in 4% of cases.95 In the Dutch cohort, 44 out of 48 patients infected with genotype 4 achieved SVR. The four failures occurred in one patient infected with subtype 4k (sofosbuvir/ledipasvir), two patients infected with subtype 4n (sofosbuvir/ledipasvir and sofosbuvir plus simeprevir) and one patient infected with subtype 4v (elbasvir/grazoprevir). The four patients infected with subtype 4r achieved SVR in this study.96
HCV genotype 5
Only one unassigned genotype 5 subtype has been identified so far, and the reported SVR rates for subtype 5a are in the same range as those for other common genotypes/subtypes.5
HCV genotype 6
Little clinical information is available on the DAA susceptibility of the many subtypes of genotype 6 circulating in Asia. Due to regional differences in their distribution and the small numbers of patients from each subtype generally included, it is difficult to draw conclusions about the true clinical susceptibility of different genotype 6 subtypes to DAA therapies.
In a recent Indian study, 53 out of 2052 patients (2.6%) with chronic HCV infection, including 81% who were HIV coinfected, were infected with genotype 6. Nine different subtypes were identified, including 6a, 6b, 6f, 6i, 6n, 6u, 6v, 6w and 6xa. Subtype 6xa was predominant (41%), followed by subtype 6n (40%). Patients were treated with a combination of sofosbuvir and daclatasvir, with or without ribavirin. The SVR rate was 81% (43/52), but the report does not specify which subtypes did not achieve SVR.101 In a real-world study from Myanmar, 39 patients infected with genotype 6 were treated with sofosbuvir/ledipasvir. 36 of them were infected with subtype 6c-l (individual subtypes not identified), 1 with 6m and 2 with unassigned subtype 6. The SVR rate was only 64% (25/39).19 16 patients infected with genotype 6 (mainly 6n) were enrolled in a clinical trial testing the combination of sofosbuvir and ravidasvir, an off-label NS5A inhibitor. The SVR rate was 81% (13/16), lower than for any other HCV genotype. Two patients with virological failure infected with subtype 6n had the double F28L+T93S NS5A RAS at baseline.92
In contrast, in a US cohort of Asian-born patients (43% of subtype 6c-l and 22% of 6a/b), SVR to sofosbuvir/ledipasvir was achieved in 95% of cases (57/60).102 Another study from Vietnam enrolled 41 patients with advanced fibrosis or cirrhosis infected with genotype 6, including 51% with 6a and 34% with 6e. The remaining patients were infected with subtypes 6k, 6h, 6l or 6o. Their SVR rate with sofosbuvir and daclatasvir was 100%.93 Other studies from different countries, including France, the USA and New Zealand, with different subtype distributions have also reported high SVR rates with different DAA regimens.81 85 99 103 104
Only 11 patients infected with genotype 6 were included in the Dutch nationwide cohort (6a, n=6; 6e, n=3; 6f, n=1; and unassigned 6, n=1). SVR was achieved in 91% of cases, with failure occurring in the patient with subtype 6f infection.96 In the European Resistance Database study, seven patients infected with genotype 6 (6e, 6f, 6n and 6r) failed to achieve SVR with ombitasvir/paritaprevir/ritonavir (two cases), sofosbuvir/ledipasvir (two cases), grazoprevir/elbasvir (one case) or sofosbuvir/velpatasvir (one case).95
HCV genotypes 7 and 8
Only anecdotal cases of patients infected with these genotypes receiving DAA treatment have been reported thus far.82
Response of unusual HCV subtypes to retreatment after DAA failure
Failure of DAA therapy to achieve SVR in patients with unusual HCV subtypes is characterised by the persistence at failure of the natural polymorphisms at RAS positions characteristic of that subtype. In addition, RASs known to confer resistance to the common genotypes are often also selected, resulting in complex resistance patterns at failure in these patients that may be difficult to retreat.
Retreatment of patients infected with unusual HCV subtypes who failed to achieve SVR after first-line therapy has been reported in several studies. The retreatment regimens were generally empirical (no protocol-based clinical trials), often but not always based on pangenotypic combinations, including sofosbuvir/velpatasvir, glecaprevir/pibrentasvir, sofosbuvir/velpatasvir/voxilaprevir or sofosbuvir plus glecaprevir/pibrentasvir. The SVR rates were high (>90%) with these combinations, but some patients failed to eliminate HCV on retreatment. However, the information is fragmentary and does not allow firm conclusions to be drawn about the resistance of some specific subtypes to retreatment.
In the London cohort of African-born patients infected with an unusual genotype 1 subtype, one patient with an unassigned genotype 1 subtype failed to respond to 16 weeks of glecaprevir/pibrentasvir.14 In the HCV Research UK cohort, eight patients infected with an unusual subtype were retreated. Two patients infected with subtype 1l and one with subtype 4r who were retreated with the same combination as first-line therapy failed to achieve SVR, as did one patient infected with subtype 4r who was retreated with sofosbuvir/velpatasvir/voxilaprevir.15
In our experience at the French National Reference Center for Viral Hepatitis B, C and D, 26 out of 43 patients infected with unusual genotype 1 subtypes were retreated. All of them achieved SVR except for one patient who was suboptimally retreated with a combination of sofosbuvir and daclatasvir. Importantly, all patients treated with a triple DAA combination or with glecaprevir/pibrentasvir achieved SVR.21 Eight patients infected with subtype 4r who failed first-line therapy achieved SVR after retreatment with a triple DAA combination in five cases and with glecaprevir/pibrentasvir in three cases.20 These results are in keeping with those of a single-arm clinical trial conducted in Rwanda in 40 patients infected with various, mostly genotype 4 unusual subtypes (4r, n=18; 4k, n=6; 4b, n=5; 4q, n=4; 4l, n=2; 4a, n=1; 4m, n=1; unassigned 4, n=1; 3h, n=1; and undetermined, n=1), all retreated with sofosbuvir/velpatasvir/voxilaprevir. The SVR rate was 98%, with only one patient infected with subtype 4q failing to achieve SVR.105
10 of the 12 patients with an unusual subtype in the Dutch cohort were retreated with different DAA combinations and all achieved SVR.96 In the European Database Study, 45 out of 48 retreated patients achieved SVR, again with different DAA combinations. The three failures were subtype 4f with glecaprevir/pibrentasvir, subtype 4r with sofosbuvir/velpatasvir and subtype 3b with sofosbuvir/velpatasvir/voxilaprevir.95
Overall, despite the paucity of data on unusual subtypes, the recommendation for retreatment after DAA failure is the same as for common subtypes, that is, a triple combination of sofosbuvir, a protease inhibitor and an NS5A inhibitor (sofosbuvir/velpatasvir/voxilaprevir or sofosbuvir plus glecaprevir/pibrentasvir). The latter should be preferred to the former in patients infected with subtype 3b (and possibly others, pending further data) because of the consistently reduced susceptibility to velpatasvir.
Conclusions
The high genetic diversity of HCV has led to the emergence of eight genotypes from a common ancestor and the subsequent diversification and spread of a large number of subtypes in restricted geographical areas. Only a small subset of these have become epidemic and are now highly prevalent on all five continents. Currently approved pangenotypic DAA regimens have been designed and developed to be effective against the most common subtypes (1a, 1b, 2a, 2b, 2c, 3a, 4a, 5a and 6a). However, large populations living in Africa and Asia, or who have migrated from these regions to industrialised countries, are infected with unusual, non-epidemic HCV subtypes, including some that are inherently resistant to currently available DAAs due to the presence of natural polymorphisms at RAS positions. Specifically, subtypes 1l, 3b, 3g, 6u and 6v have been identified as resistant to most NS5A inhibitors, with pibrentasvir demonstrating better efficacy than other members of the drug class. Other assigned or yet unassigned HCV subtypes may also have reduced DAA susceptibility, but the data are limited. The high prevalence of these subtypes in low-income and middle-income countries, where only older-generation DAAs are available, particularly the generic combinations of sofosbuvir and daclatasvir or sofosbuvir/ledipasvir, challenges the WHO HCV elimination targets in these regions. Failures have also been reported with the most recent pangenotypic double combinations, including sofosbuvir/velpatasvir and glecaprevir/pibrentasvir.
In 2020, EASL recommended that, in settings where sequencing of the NS5A region by means of population or deep sequencing is available and affordable, patients infected with subtypes 1l, 3b, 3g, 6u and 6v and those infected with other unusual subtypes harbouring more than one RAS known to confer resistance to NS5A inhibitors should be considered for first-line treatment with the triple combination of sofosbuvir, velpatasvir and voxilaprevir.13 This recommendation is followed in our hepatology centre, where every newly diagnosed HCV patient who was born in Africa or Asia benefits from NS5A sequencing and tailored therapy. However, in the era of treatment simplification and large-scale elimination, this recommendation can only be applied in a very limited number of highly specialised tertiary referral centres, while the vast majority of patients are treated in other settings. Figure 6 shows a treatment decision tree for patients with HCV infection born in Africa or Asia according to the availability/affordability of NS5A sequencing.
Treatment decision tree in patients with HCV infection without cirrhosis or with compensated cirrhosis born in Africa or Asia according to the availability/affordability of NS5A sequencing. HCV, hepatitis C virus; SVR, sustained virological response; RAS, resistance-associated substitution; SOF, sofosbuvir; VEL, velpatasvir; VOX, voxilaprevir; G, glecaprevir; P, pibrentasvir; RBV, ribavirin; wk, week. *According to the EASL 2020 Recommendations on Treatment of Hepatitis C.13
Several options exist to minimise the impact of unusual subtypes that are inherently resistant to DAA regimens on HCV elimination at the country level. (1) It is essential to characterise the distribution of viral subtypes circulating in each country, especially in low-income and middle-income countries where this information is currently lacking, in order to characterise the prevalence of natural polymorphisms that may influence response to antiviral treatment and to tailor first-line therapy to the true epidemiology of HCV in the country. Regular updates are also needed in industrialised countries where most newly diagnosed cases are now immigrants from the same regions. Since routine sequencing is not feasible on a large scale, sofosbuvir/velpatasvir/voxilaprevir or sofosbuvir plus glecaprevir/pibrentasvir must be made available as a cheap, generic first-line therapy wherever inherently resistant HCV subtypes are prevalent. (2) If the triple combination is not available or affordable as first-line therapy in areas with a large prevalence of HCV subtypes that are inherently resistant to DAAs, SVR at 12 weeks post-treatment must be systematically monitored, and patients who fail must be retreated with a triple combination as second-line therapy. If this is not done, a substantial proportion of individuals will fail to clear the virus and may transmit it to others.
Overall, the problem of unusual HCV subtypes that are inherently resistant to DAAs and its threat to the global efforts to eliminate viral hepatitis are underestimated and warrant vigorous action.
Ethics statements
Patient consent for publication
Ethics approval
Not applicable.
Acknowledgments
The authors are grateful to Ms Melissa N’Debi for generating the phylogenetic tree presented in Figure 1.
References
Supplementary materials
Supplementary Data
This web only file has been produced by the BMJ Publishing Group from an electronic file supplied by the author(s) and has not been edited for content.
Footnotes
Contributors EV-Q: conceptualisation, methodology, literature search, and writing of the original draft and editing of revised versions. J-MP: conceptualisation, methodology, literature search, writing and editing of the revised versions and supervision.
Funding The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Map disclaimer The inclusion of any map (including the depiction of any boundaries therein), or of any geographic or locational reference, does not imply the expression of any opinion whatsoever on the part of BMJ concerning the legal status of any country, territory, jurisdiction or area or of its authorities. Any such expression remains solely that of the relevant source and is not endorsed by BMJ. Maps are provided without any warranty of any kind, either expressed or implied.
Competing interests EV-Q has no conflict of interest to disclose. J-MP has served as an advisor or speaker for Abbott, AbbVie, Gilead and GSK.
Provenance and peer review Not commissioned; externally peer reviewed.
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.