Article Text
Statistics from Altmetric.com
We read with great interest the paper by Ha et al 1 demonstrating that circulating levels of serotonin (5-hydroxytryptamine, 5-HT) are increased in COVID-19 and correlate with disease severity and gastrointestinal symptoms such as diarrhoea. Another recent paper by Lin et al 2 in this journal demonstrated that diarrhoea is the most common GI symptom in patients with COVID-19. Almost all 5-HT in our body is produced by enterochromaffin (EC) cells within the epithelium of the GI tract, which constitute approximately half of all enteroendocrine (EE) cells. Gut-derived 5-HT modulates gut peristalsis and exacerbates inflammatory responses by acting as a chemotactic molecule for various immune cells and by triggering cytokine release.3 While most gut epithelial cell types are susceptible to SARS-CoV-2 infection, EE cells have the greatest proportion of cells infected at 12 hours after viral exposure.4 In addition, the use of selective serotonin reuptake inhibitors (SSRI), normally prescribed to treat mental health conditions such as depression, is reported to reduce COVID-19 severity in humans.5 The GI tract is a route of SARS-CoV-2;6 however, its unknown if EC cells have any specific capacity for infection that would explain the increased 5-HT in patients with COVID-19, or the SSRI treatment efficacy reported. We, therefore, examined (see online supplemental file 1) the transcriptomes of cells lining the gut wall7 for expression of genes associated with SARS-CoV-2 infection, with a focus on EE cell subtypes.
Supplemental material
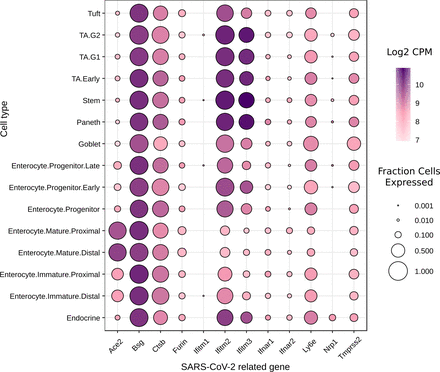
Our focus was on gene expression for proteins implicated or known to be involved as COVID-19 receptors for efficient cell entry; ACE2, BSG and NRP1, associated proteins involved in intracellular trafficking and breakdown; TMPRSS2, FURIN and CTSB, and proteins associated with viral protection; LY6E, IFITM1-3 and IFNAR1-2. We identified that the genes encoding for all of these proteins are expressed within the intestinal epithelium (figure 1). Of the known COVID-19 receptors, Ace2 and Bsg genes are highly expressed in all epithelial cell types. However, the more recently identified receptor, NRP1, is expressed exclusively in hormone-producing EE cells at the gene level.
Absolute expression of SARS-CoV-2-related genes in different cell types from the mouse small intestinal epithelium.7 Data are shown as colour scale of Log2 of mean copies per million (CPM), with circle size indicating fraction of each cell type expressing the gene.
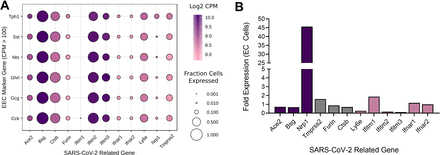
To examine this further, we determined which subtypes of EE cells express Nrp1 (figure 2). We focused on the major EE cell types containing cholecystokinin, glucagon-like peptide-1, ghrelin, neurotensin, somatostatin and 5-HT (figure 2A). EC cells that express tryptophan hydroxylase 1 (Tph1), the rate-limiting enzyme for non-neuronal 5-HT synthesis, are the primary cell type in the gut wall expressing Nrp1, indicating these cells may be a route of infection and disease pathogenesis. We then focused solely on EC cell gene expression using a second RNA-seq database8 and found that while all COVID-19-related genes are expressed in EC cells, Nrp1 has the greatest enrichment of expression of all these, of approximately 45-fold greater expression in EC cells than in non-EC epithelial cells (figure 2B). Subsequent examination of published work identifies that NRP1 protein expression is highly colocalised in the gastrointestinal wall with cells that express chromogranin-A, a marker of EC cells.9
Absolute expression levels of SARS-CoV-2 related genes in different enteroendocrine cell (EEC) types from the mouse small intestinal epithelium.7 Data are shown as colour scale of Log2 of mean copies per million (CPM), with circle size indicating fraction of each cell type expressing the gene.
Cells expressing NRP1, ACE2 and TMPRSS2 have a >3-fold increase in SARS-CoV-2 infection compared with ACE2 and TMPRSS2 alone.10 Our data demonstrate that EC cells are the only gut cell type that expresses significant levels of these three SARS-CoV-2-related genes. This, therefore, provides a link between EC cells and the increased diarrhoea,2 circulating 5-HT,1 and efficacy of SSRIs5 that are reported in COVID-19. Experiments investigating SARS-CoV-2 infectivity in the absence of gut-derived 5-HT would provide further evidence of this link.
Ethics statements
Patient consent for publication
Ethics approval
Not applicable.
Supplementary materials
Supplementary Data
This web only file has been produced by the BMJ Publishing Group from an electronic file supplied by the author(s) and has not been edited for content.
Footnotes
Twitter @alyce_martin, @ConfocalJones
Contributors AMM, LAJ and DJK developed the concept; MR and RE performed bioinformatic analysis; MR and AMM made the figures; AMM, LAJ, DT, RAC, CA and DJK reviewed the literature and wrote the drafts; all authors read and approved the final manuscript for submission.
Funding Supported by grants from the Australian Research Council (DE220100403 to AMM).
Competing interests None declared.
Provenance and peer review Not commissioned; internally peer reviewed.
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.